The TRANSFAC project
| The "huge" number of eukaryotic transcription factors (TFs) prompted me to make some early attempts to review and systematize them1, 2. At that time, although the number of newly discovered TFs seemed to grow rapidly and getting unmanageable, we know now that it was a tiny percentage of what we know today, about 20 years later. Also the number of TF binding sites (TFBSs) in promoters and enhancers grew quickly, so that I decided to put the available knowledge about transcriptional regulators and their target sequences together in one compilation3. That compilation anticipated already the later core structure of the TRANSFAC database in arranging the feature of binding sites and the binding TFs in two separate tables, referencing to each other by the names of the TFs. A third table compiled the amino acid sequences of the one motif mediating sequence-specific DNA-binding that was known at that time for eukaryotic TFs, the zinc finger. |  | 
|
The front page and one table page of the original compilation3, which laid the foundations for the later TRANSFAC database. |
||
The whole project was started as a purely private initiative, on my own ATARI computer. A smart student of our department, Thomas Heinemeyer, stepped in and migrated the binding site data from a simple text file into a reference database, which had to be tweeked quite a bit to deal with this type of data. I did it then with the factor data, and used the system for maintaining the database for quite a while. With the head of our department, John Collins, we found a dedicated well-wisher of this little initiative since he recognized its potential value for the upcoming human genome research.
With the advent of the first bioinformatics funding program in 1993, I got the chance to render the TRANSFAC initiative a real scientific project as part of the GENUS consortium. Now, we made TRANSFAC a relational database, with the tables FACTOR and SITE as core components, the link between both tables representing the protein-DNA interaction and having a score attributed that reflected the level of experimental validation.
We had recognized the variability among all sites one TF can interact with, and had already suggested first algorithms how to cope with this fact with regard to characterizing and predicting these sites way before this project started4. But now that we had sufficient experimental data at hand and well-structured documented in the TRANSFAC database, we could start mining these data. At least for some TFs where a sufficient number of binding sites was published, we aligned their binding sites and established a library of position scoring matrices that quickly became the "fillet" of TRANSFAC. The program MatInspector5 that was developed in a joint project with Thomas Werner's group made use of this library in predicting potential TFBSs and became soon a very popular tool.
Finally, the central "axis" of TRANSFAC made up by experimental sites and factors and their link/interaction was accompanied by two kinds of abstractions: binding sites of one factor were abstracted to a positional scoring matrix, and transcription factors were grouped into classes according to the features of their DNA-binding domains. This developed further into a more elaborate trancription factor classification (see the corresponding project description)6.
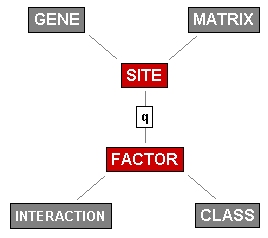 | The early conceptual schema of TRANSFAC: The central axis between the tables SITE and FACTOR represents the DNA-protein interactions at regulatory elements of a GENE, to which each element can be assigned to. The individual DNA-protein interactions are qualified by a parameter q according to their experimental validation. Most transcription factors exert a number of mutual protein-protein interactions, which have been defined in the table INTERACTION. In contrast to the other tables, MATRIX and CLASS do not document primary experimental information, but provide abstractions by summarizing a number of sites in a positional scoring matrix or classifying transcription factors according to their DNA-binding domains. |
After about 10 years of development, we made TRANSFAC the core product of a new company, BIOBASE, in order to ensure sustainable income for its survival, after it has become an important part of the international bioinformatics infrastructure. TRANSFAC® is now a registered trademark of BIOBASE. It is accompanied a TFBS prediction tool, MATCHTM (as acronym for MATrix searCH), which still applies most of the principles of the early algorithm7 and is a trademark of BIOBASE. An up-to-date report on the status of TRANSFAC and the accompanying tool is given in Wingender, 20088.
A free version can be accessed here (registration required). Further information is provided by Wikipedia.
Selected publications:
1 Wingender, E. and Seifart, K. H.: Transcription in eukaryotes - the role of transcription complexes and their components. Angew. Chem. Int. Ed. Engl. 26, 218-227 (1987). (PubMed: )
2 Wingender, E.: Transcription regulating proteins and their recognition sequences. CRC Crit. Rev. in Eukaryotic Gene Expression 1, 11-48 (1990). (PubMed: )
3 Wingender, E.: Compilation of transcription regulating proteins. Nucleic Acids Res. 16, 1879-1902 (1988). (PubMed: 3282223)
4 Wingender, E., Heinemeyer, T. and Lincoln, D.: Regulatory DNA sequences: predictability of their function. Genome Analysis - From Sequence to Function; BioTechForum- Advances in Molecular Genetics (J. Collins, A.J. Driesel, eds.) 4, 95-108 (1991). (pdf)
5 Quandt, K., Frech, K., Karas, H., Wingender, E. and Werner, T.: MatInd and MatInspector - New fast and sensitive tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23, 4878-4884 (1995). (PubMed: 8532532)
6 Wingender, E.: Classification scheme of eukaryotic transcription factors. Mol. Biol. (Mosk.) 31, 584-600 (1997); Mol. Biol. Engl. Tr. 31, 483-497 (1997). (PubMed: 9340487)
7 Kel, A. E., Gößling, E., Reuter, I., Cheremushkin, E., Kel-Margoulis, O. V. and Wingender, E.: MATCHTM: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 31, 3576-3579 (2003). (PubMed: 12824369)
8 Wingender, E.: The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief. Bioinform. 9, 326-332 (2008). (PubMed: 18436575)