Completed projects
Altogether, I have been involved so far in 9 major projects funded by the German Ministry of Education and Research (BMBF) and 6 project funded by the European Commission. On this page, or linked to it, I will add by and by the description of all these projects. The time line of the externally funded projects is like this:
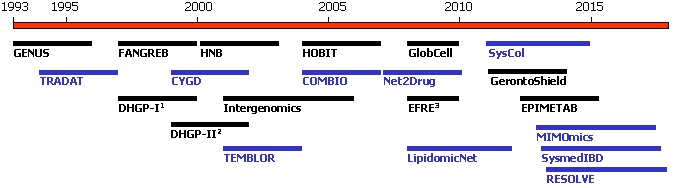
The projects funded by the German Ministry of Education and Research are indicated by a black, those supported by the European Union by a blue bar.
1, name of the funding program given; the full title of the project was "Computer-aided automatic sequence analysis of the human genome"
2, name of the funding program given; the full title of the project was "Modeling of gene regulatory networks for linking genotype-phenotype information"
3, name of the funding program given; the full title of the project is "Molecular representation of disease states in biological regulatory networks"
Besides, a number of small-scale international collaboration projects with partners in Russia, China and Japan could be accomplished thanks to additional funding by the International Bureau of the German Research Ministry.
EFRE (2009 – 2011)
This project was funded by the the Land Niedersachsen and the European Regional Development Fund (ERDF, German acronym: EFRE). It aimed at developing approaches and tools for the computer-assisted representation and modeling of various cellular states associated with diseases. The underlying idea was that a specific disease state is a particular cellular state, maybe even as stable as any normal cellular state, so that it should be possible to characterize it as a specific activity state of the regulatory network of the affected cell(s). After a prolongation, the project ended March, 2011.
You may learn more about the UMG activities in this project here.
The partner of this project was BIOBASE GmbH, Wolfenbüttel, Germany
LipidomicNet (2008 – 2012)


LipidomicNet was a large EU-funded project where my department at UMG is contributing to the bioinformatics tasks. Our main activities were focussing on the extension of the EndoNet resource towards the systemic regulation of the lipid metabolism. The goal was to provide a (static) model of the regulatory network between the most relevant organs, mainly liver, pancreas, and adipose tissue. While my department at UMG dealt with the intercellular pathways, the corresponding processes inside the connected cells were modeled by the TRANSPATH database of BIOBASE, which was another partner in this consortium. Both data resources were integrated under the BioUML platform of ISB (Institute of Systems Biology in Novosibirsk), also a consortial partner.
One of the major resource for EndoNet is the Cytomer ontology of human anatomical structures, tissues and cell types at different developmental stages. To handle this and other ontologies, and make them easily accessible from other applications, a dedicated OBA server was developed.1
This project ended by 31st of October, 2012. The closing project meeting was held in Regensburg, Germany, on October 8th-10th, 2012.
You may learn more about the UMG contribution to LipidomicNet here or about the consortium at the central LipidomicNet web pages.
Selected publications:
1 Dönitz, J. and Wingender, E.: The ontology-based answers (OBA) service: a connector for embedded usage of ontologies in applications. Front Genet. 3, 197 (2012). (PubMed: 23060901)
GlobCell (2008 – 2010)


GlobCell was an international research project, funded by the Euro Trans-Bio initiative (ETB), granted by the German Ministry of Education and Research, and coordinated by Dr. Alexander Kel at BIOBASE (now CSO of geneXplain GmbH). It aimed at analyzing and ultimately predicting the behaviour of human cells in a complex environment, using a combination of cell biology, bioinformatics, mathematical modeling and systems biology approaches and tools. After a prolongation, the project ended by December 2010.
The contribution of my department at UMG was to provide integrated semantic and mechanistic network models and methods for comparative analysis of the cellular pathways triggered by more than one stimulus.
You may learn more about the UMG contribution to GlobCell here.
Net2Drug (2007 – 2010)


Net2Drug was an international research project, funded by the EU and coordinated by Dr. Alexander Kel at BIOBASE (now CSO of geneXplain GmbH). It aimed at developing a set of tools that combines different experimental high-throughput methods together with bio- and chemoinformatical approaches.
The contribution of my department at UMG was (1) to provide an all-genome prediction of potential transcription factor binding sites (TFBSs) based on statistical scoring plus conservativity analysis across multiple genomes, and (2) to provide an integrated regulatory & metabolic network and optimal filters when mapping experimental gene expression data on such a network.
This project ended by 31st of July, 2010. The final project meeting was held in Alghero, Sardinia, on July 21st/22nd, 2010.
You may learn more about the UMG contribution to Net2Drug here or about the consortium at the central Net2Drug web pages.
COMBIO (2004 – 2007)

The full name of this project was "An integrative approach to cellular signalling and control processes: Bringing computational biology to the bench" and was coordinated by Luis Serrano, first at EMBL (Heidelberg), later at the CRG in Barcelona. Its aim was to bring together experts in modeling and simulation with experimentalists. Two systems were under study, both of which of considerable importance for the regulation of cell cycle progression: the p53-Mdm2 circuit and the formation of the mitotic spindle apparatus.
The contribution of my department at UMG mainly comprised the systematic analysis of the topological features of the extended p53 network. For this, the main methods summarized under the heading "Network analysis" were developed and applied1. For instance, the concept of the pairwise disconnectivity index was developed and applied to individual nodes, edges, and network patterns (subgraphs)2, 3. In addition, concepts were developed how to integrate pathway-related data to provide an information resource for what we may call "systems pathology"4.
The Contract number for this project was LSHG-CT-2004-503568. For further details, you may wish to go to the original COMBIO project page.
Publications:
1 Potapov, A. P., Voss, N., Sasse, N. and Wingender, E.: Topology of mammalian transcription networks. Genome Inf. Ser. 16, 270-278 (2005). (PubMed: 16901109)
2 Potapov, A. P., Goemann, B. and Wingender, E.: The pairwise disconnectivity index as a new metric for the topological analysis of regulatory networks. BMC Bioinformatics 9, 227 (2008). (PubMed: 18454847)
3 Goemann, B., Wingender, E. and Potapov, A. P.: An approach to evaluate the topological significance of motifs and other patterns in regulatory networks. BMC Syst. Biol. 3, 53 (2009). (PubMed: 19454001)
4 Wingender, E., Hogan, J., Schacherer, F., Potapov, A. P. and Kel-Margoulis, O.: Integrating pathway data for systems pathology. In Silico Biol. 7 S1, 03 (2007). (PubMed: 17822386; Link)
HOBIT (2004 – 2007)
Intergenomics (2001 – 2006)


This was by far the largest project I ever initiated and coordinated. It was one of the six Bioinformatics Competence Centers funded by the German Ministry of Education and Research, which formed the NBCC network across Germany. In the initial phase, three institutions (GBF – Gesellschaft für Biotechnologische Forschung in Braunschweig, today: Helmholtz Center for Infection Research; Technical University of Braunschweig; The University of Applied Sciences / Fachhochschule Braunschweig / Wolfenbüttel) and one company (BIOBASE GmbH in Wolfenbüttel) with altogether 11 projects worked together on the central subject: How to model the genome-driven processes during the process of bacterial infections. That was also what the name Intergenomics was indicating for: Interaction between genomes.
Because of its considerable size, it is not possible to describe here all the scientific initiatives launched in this project here. To learn more about the Intergenomics Center, you may wish to go to the Intergenomics web site of this project, which is still available. The work of my own group, which started at GBF and moved to UMG in November 2001, focussed on developing new approaches for the analysis of promoter features. The two brilliant PhD students being funded by the Center, Ekaterina Shelest and Tilman Sauer, have done excellent work on the combinatorics of different TFBS (Katya)1, 2 and the phylogenetic footprinting as additional independent criterion for single site detection3. Besides, there were nice synergies with other groups in the Center. So, based on our experiences with the TRANSFAC database, we could give some initial advice on building a similar database (PRODORIC) about prokaryotic transcription factors and their DNA-binding sites and behavior4. But also our own databases have made great progress due to the joint work of, mainly, my UMG group and BIOBASE5-8. On the algorithmic side, Alexander Kel at BIOBASE and his team together with colleagues in Novosibirsk could develop the Composite Module Analyst (CMA)9, on top of which an algorithm was developed that identifies key nodes in the signaling networks that appear to control the transcription factors of the identified composite modules10. Related with this, the DEEP algorithm was developed at UMG and shown to identify relevant subnetworks from a given gene set11.
Among the long-standing effects of this center, in my personal view, I would mainly highlight the fact that it was the essential kick to develop a real bioinformatics community in Braunschweig, a city that is rightly proud of being a science center in Germany. Before Intergenomics, however, there were a few individuals and groups that had some interest in that field, but very little active research was done. Intergenomics helped establish a new bioinformatics professorship at TU Braunschweig, and the university was lucky enough to gain Dietmar Schomburg for this position, who was coordinator of another Bioinformatics Center, CUBIC in Cologne. He is now heading the department of bioinformatics and biochemistry in the institue of biochemistry.
The final report of the Center from January 2007 can be found here (with the financial details omitted).
Publications:
1 Shelest, E., Kel, A.. E., Gößling, E. and Wingender, E.: Prediction of potential C/EBP/NF-kB composite elements using the matrix-based search methods. In Silico Biol. 2, 0007 (2003). (Link)
2 Shelest, E. and Wingender, E.: Construction of predictive promoter models on the example of antibacterial response of human epithelial cells. Theor. Biol. Med. Model. 2, 2 (2005). (PubMed: 15647113)
3 Sauer, T., Shelest, E. and Wingender, E.: Evaluating phylogenetic footprinting for human-rodent comparisons. Bioinformatics 22, 430-437 (2006). (PubMed: 16332706)
4 Münch, R., Hiller, K., Barg, H., Heldt, D., Linz, S., Wingender, E. and Jahn, D.: PRODORIC: Prokaryotic Database Of Gene Regulation. Nucleic Acids Res. 31, 266-269 (2003). (PubMed: 12519998)
5 Wingender, E.: TRANSFAC®, TRANSPATH® and CYTOMER® as starting points for an ontology of regulatory networks. In Silico Biology 4, 0006 (2003). (Link)
6 Choi, C., Crass, T., Kel, A., Kel-Margoulis, O., Krull, M., Pistor, S., Potapov, A., Voss, N., and Wingender, E.: Consistent re-modeling of signaling pathways and its implementation in the TRANSPATH database. Genome Inf. Ser. 15, 244-254 (2004). (PubMed: 15706510)
7 Krull, M., Pistor, S., Voss, N., Kel, A., Reuter, I., Kronenberg, D., Michael, H., Schwarzer, K., Potapov, A., Choi, C., Kel-Margoulis, O. and Wingender, E.: TRANSPATH®: an Information resource for storing and visualizing signaling pathways and their pathological aberrations. Nucleic Acids Res. 34, D546-D551 (2006). (PubMed: 12519957)
8 Matys, V., Kel-Margoulis, O. V., Fricke, E., Liebich, I., Land, S., Barre-Dirrie, A., Reuter, I., Chekmenev, D., Krull, M., Hornischer, K., Voss, N., Stegmaier, P., Lewicki-Potapov, B., Saxel, H., Kel, A. E. and Wingender, E.: TRANSFAC® and its module TRANSCompel®: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108-D110 (2006). (PubMed: 16381825)
9Kel, A., Konovalova, T., Waleev, T., Cheremushkin, E., Kel-Margoulis, O. and Wingender, E.: Composite Module Analyst: a fitness-based tool for identification of transcription factor binding site combinations. Bioinformatics 22, 1190-1197 (2006). (PubMed: 16473870)
10Kel, A., Voss, N., Jauregui, R., Kel-Margoulis, O. and Wingender, E.: Beyond microarrays: Find key transcription factors controlling signal transduction pathways. BMC Bioinformatics 7 Suppl. 2, S13 (2006). (PubMed: 17118134)
11Degenhardt, J., Haubrock, M., Wingender, E. and Crass, T.: DEEP - A Tool for Differential Expression Effector Prediction. Nucleic Acids Res. 35, W619-W624 (2007). (PubMed: 17584786)
TEMBLOR (2002 – 2005)

Initially called "Integr8", this project was launched under coordination of the European Bioinformatics Institute (EBI) to faclitate the interoperability of a wide range of European databases (EU project number: QLRI-CT-2001-00015).
The UMG contribution to this project was to develop further our set of databases and facilitate their integration. In this context, mainly our databases S/MARt DB (about scaffold / matrix attached regions) and ReAlSplice (about regulated alterntative splicing events) were incorporated.
HNB (2000 – 2005)



The Helmholtz Network for Bioinformatics (HNB), originally planned as "Virtual Bioinformatics Center" in Germany, was a joint venture of 12 German bioinformatics research groups. Its aim was to offer convenient access to numerous bioinformatics resources through a single web portal. The task of our group was to provide the central web resources.
The HNB was funded by the German Ministry of Education and Research (BMBF, 01SF9988/4) and was supported by the Helmholtz Association of German Research Centres (HGF). More information can be found here. A poster on the HNB tasks was presented at the ECCB 2002. The project was continued with a shifted focus as "HOBIT" project.
Publications:
Crass, T., Antes, I., Basekow, R., Bork, P., Buning, C., Christensen, M., Claußen, H., Ebeling, C., Ernst, P., Gailus-Durner, V., Glatting, K.-H., Gohla, R., Gößling, F., Grote, K., Heidke, K., Herrmann, A., O'Keeffe, S., Kießlich, O., Kolibal, S., Korbel, J. O., Lengauer, T., Liebich, I., van der Linden, M., Luz, H., Meissner, K., von Mering, C., Mevissen, H.-T., Mewes, H.-W., Michael, H., Mokrejs, M., Müller, T., Pospisil, H., Rarey, M., Reich, J. G., Schneider, R., Schomburg, D., Schulze-Kremer, S., Schwarzer, K., Sommer, I., Springstubbe, S., Suhai, S., Thoppae, G., Vingron, M., Warfsmann, J., Werner, T., Wetzler, D., Wingender, E. and Zimmer, R.: The Helmholtz Network for Bioinformatics: an integrative web portal for bioinformatics resources. Bioinformatics 19, 268-270 (2004). (PubMed: 14734319)
DHGP-II (2000 – 2004)

The full title of this consortial research project, which I had the honor to coordinate, was "Modeling of gene regulatory networks for linking genotype-phenotype information". It received funding from the German Human Genome Program II (funding code 01KW 9912). I started with the project while still at GBF, but moved with the two scientists working on the project to Goettingen University by end of 2001. The project partners were Thomas Werner (GSF), Dietmar Schomburg (University of Cologne), Ralf Hofestaedt (University of Magdeburg), and Friedrich-Karl Trefz (University of Tübingen, Clinics in Reutlingen).
It was the aim of this project to develop bioinformatics tools for investigating genetic mutations which exhibit clinically relevant phenotypes. That included developing, adapting and integrating data sources about transcriptional regulation, signal transduction pathways, metabolic pathways, cellular systems, and metabolic diseases with programs for the prediction of regulatory genome regions and simulation of regulatory pathways. It also aimed at creating an integrative system that provides a unifying view of molecular mechanisms in gene regulation, signal transduction, metabolic pathways, and clinical consequences of their aberrations. As a case study, the focus was put on certain types of diabetes (MODY) where disregulation of transcriptional control leads to the aberrant metabolic phenomena that are characteristic for diabetes.
Among our achievements in this project were the first models for regulatory pathways1, 2. Some of the newly developed concepts found their way into the data model of the TRANSPATH database3, and the forced data acquisition for insulin pathways and related signaling cascades enhanced the contents of TRANSPATH4, 5.
Publications:
1 Potapov, A. and Wingender, E.: Modeling the architecture of regulatory networks. Computer Science and Biology. Proceedings of the German Conference on Bioinformatics (GCB 2001). E. Wingender, R. Hofestädt, I. Liebich (eds.). GBF-Braunschweig, pp. 6-10 (2001). (link)
2 Potapov, A. P. and Wingender, E.: Representing the architecture of signal transduction networks in an algebraic form: protein target finding. Ann. N. Y. Acad. Sci. 973, 1-2 (2002). (PubMed: 12485823)
3 Choi, C., Crass, T., Kel, A., Kel-Margoulis, O., Krull, M., Pistor, S., Potapov, A., Voss, N., and Wingender, E.: Consistent re-modeling of signaling pathways and its implementation in the TRANSPATH database. Genome Inf. Ser. 15, 244-254 (2004). (PubMed: 15706510)
4 Krull, M., Voss, N., Choi, C., Pistor, S., Potapov, A. and Wingender, E.: TRANSPATH®: an integrated database on signal transduction and a tool for array analysis. Nucleic Acids Res. 31, 97-100 (2003). (PubMed: 12519957)
5 Choi, C., Krull, M., Kel-Magoulis, O., Pistor, S., Potapov, A., Voss, N. and Wingender, E.: TRANSPATH® - a high quality database on signal transduction. Comparative Functional Genomics 5, 163-168 (2004). (PubMed: 18629064)
CYGD (2000 – 2003)

CYGD was a EU-funded project under coordination of H.-W. Mewes (MIPS-GSF, Martinsried, Germany; project number was QLRI-1999-01333). Its aim was to build a comprehensive database incorporating genome-related information about yeast beyond mere sequence data. Our contribution, achieved by Holger Michael, was to bring in an updated TRANSFAC data set of yeast transcription factors, their genomic binding sites and DNA-bindig profiles. These data were integrated into the CYGD platform, but also constituted an independent module "TSM" (TRANSFAC Saccharomyces Module) which has been made freely available.
Publications:
1 Güldener, U., Münsterkötter, M., Kastenmüller, G., Strack, N., van Helden, J., Lemer, C., Richelles, J., Wodak, S. J., García-Martínez, J., Pérez-Ortín, J. E., Michael, H., Kaps, A., Talla, E., Dujon, B., André, B., Souciet, J. L., De Montigny, J., Bon, E., Gaillardin, C. and Mewes, H. W.: CYGD: the Comprehensive Yeast Genome Database. Nucleic Acids Res. 33, D364-D368 (2005). (PubMed: 15608217)
DHGP-I (1997 – 2000)

We had a minor contribution to this consortial research project, which was funded in the framework of the first German Human Genome Project. The consortial task was to develop methods for "Computer-assisted editing of genomic sequences", coordinated by Sandor Suhai at the German Cancer Research Center (DKFZ), Heidelberg; our project part was entitled "Computer-aided automatic sequence analysis of the human genome" (funding code: 01KW9629/7). In this project, we enforced the biological network idea by expanding the TRANSFAC concept towards those signaling pathways which regulate the activity of transcription factors. Our attempts to design, implement and populate a database on these pathways then prompted Frank Schacherer in my group to develop the TRANSPATH database with its multiple abstraction layers. Together with the Cell Signaling Network Database (CSNDB) of Takako Takai-Igarashi, Tokyo, from whom we received significant help in the early stages of our own work, these were the first two signaling pathway databases ever developed and made available to the scientific community.
Today, TRANSPATH® is maintained and distributed by BIOBASE.
Publications:
1 Heinemeyer, T., Chen, X., Karas, H., Kel, A. E., Kel, O. V., Liebich, I., Meinhardt, T., Reuter, I., Schacherer, F. and Wingender, E.: Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 27, 318-322 (1999). (PubMed: 9847216)
2 Wingender, E., Chen, X., Hehl, R., Karas, H., Liebich, I., Matys, V., Meinhardt, T., Prüß, M., Reuter, I. and Schacherer, F.: TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 28, 316-319 (2000). (PubMed: 10592259)
3 Wingender, E., Chen, X., Fricke, E., Geffers, R., Hehl, R., Liebich, I., Krull, M., Matys, V., Michael, H., Ohnhäuser, R., Prüß, M., Schacherer, F., Thiele, S. and Urbach, S.: The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 29, 281-283 (2001). (PubMed: 11125113)
4 Schacherer, F., Choi, C., Götze, U., Krull, M., Pistor, S. and Wingender, E.: The TRANSPATH signal transduction database: a knowledge base on signal transduction networks. Bioinformatics 17, 1053-1057 (2001). (PubMed: 11724734)
FANGREB (1997 – 2000)

Essentially, this project was mainly a continuation of the GENUS project. The full title was "Functional annotation of regulatory genome regions" (with FANGREB being the acronym of the German title "Funktionelle Annotation regulatorischer Genombereiche"; funding number 0311640), and I had the honor to coordinate this project. From the very beginning of this project, the responsible people in the ministry made clear to us that if we wish to continue with providing and further maintaining our resources, which (like the TRANSFAC database and the MatInspector tool) became already appreciated parts of the international research infrastructure, we have to find a mechanism that ensure self-sustainement. That was when our partner Thomas Werner decided to establish Genomatix, and we founded BIOBASE.
Scientifially, we succeeded to develop the TRANSFAC database1, 2 significantly further. At that time, TRANSFAC could also be complemented by a database on composite elements, COMPEL3. The latter was a collaborative effort together with our colleagues at the Institute of Cytology and Genetics in Novosibirsk, Russia, where Olga Kel-Margoulis did an extremely good job in giving this type of data a standardized structure with the advice from Prof. A. Romashchenko and Prof. Nikolay Kolchanov. Building on the insights obtained from this data collection, we could establish an efficient algorithm to reliably detect one type of composite elements (NFAT/AP-1)4; that was mainly the work of Alexander Kel.
Also out of the FANGREB project, we could launch another international collaboration; a visiting scientist and PhD student of mine, Xin Chen from Peking University, Beijing, China, joined my group and developed a nice ontology on cell types, tissues, organs, which we called CYTOMER1, 5. It was thought as a framework to map gene expression patterns, first for human, later for mouse systems as well.
The final summarizing report of the whole consortial project from November 2000 can be found here, and the more detailed report of our subproject here (in German only).
Publications:
1 Heinemeyer, T., Chen, X., Karas, H., Kel, A. E., Kel, O. V., Liebich, I., Meinhardt, T., Reuter, I., Schacherer, F. and Wingender, E.: Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 27, 318-322 (1999). (PubMed: 9847216)
2 Wingender, E., Chen, X., Hehl, R., Karas, H., Liebich, I., Matys, V., Meinhardt, T., Prüß, M., Reuter, I. and Schacherer, F.: TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 28, 316-319 (2000). (PubMed: 10592259)
3 Kel-Margoulis, O. V., Romashchenko, A. G., Kolchanov, N. A., Wingender, E. and Kel, A. E.: COMPEL: a database on composite regulatory elements providing combinatorial transcriptional regulation. Nucleic Acids Res. 28, 311-315 (2000). (PubMed: 10592258)
4 Kel, A., Kel-Margoulis, O., Babenko, V. and Wingender, E.: Recognition of NFATp/AP-1 composite elements within genes induced upon the activation of immune cells. J. Mol. Biol. 288, 353-376 (1999). (PubMed: 10329147)
5 Chen, X., Dress, A., Karas, H., Reuter, I. and Wingender, E.: A database framework for mapping expression patterns. Computer Science and Biology. Proceedings of the German Conference on Bioinformatics (GCB '99). R. Giegerich, R. Hofestädt, T. Lengauer, W. Mewes, D. Schomburg, M. Vingron and E. Wingender (eds.). GBF-Braunschweig and University of Bielefeld, pp. 174-178 (1999). (Link)
TRADAT (1995 – 1998)

The full title of this project, which I was the coordinator of, was "Characterization of regulatory genomic regions". It received funding from the European Union (BIO4-CT95-0226). Besides my own institue (GBF), the partnership comprised the groups of Luciano Milanesi (ITBA-CNR, Milan, Italy), Martin J. Bishop (MRC, Cambridge, UK), Thomas Werner (GSF), Philipp Bucher (ISREC, Epalinges, Switzerland), and Nicolas Mermod (Lausanne, Switzerland).
The objectives of the project were to develop and provide tools for the interpretation of genomic DNA sequences with special emphasis on regulatory regions. During this project, both the Eukaryotic Promoter Database (EPD) as ISREC as well as our TRANSFAC database1, 2, 3 gained a lot of contents and functionality. This project enabled also to establish the first transcription factor classification4. The matrix-based approach to detect TFBSs was complemented by a method to define proper acceptance threshold, and to do so in a context-sensitive manner with help of a fuzzy clustering method5.
The final report can be downloaded, text and figures separately.
Publications:
1 Wingender, E., Karas, H. and Knüppel, R.: TRANSFAC Database as a Bridge between Sequence Data Libraries and Biological Function. Pacific Symposium on Biocomputing '97 (PSB'97), R. B. Altman, A. K. Dunker, L. Hunter, T. E. Klein (eds.). World Scientific, Singapore - New Jersey - London - Hong Kong 1996, pp. 477-485. (PubMed: 9390316)
2 Heinemeyer, T., Wingender, E., Reuter, I., Hermjakob, H., Kel, A. E., Kel, O. V., Ignatieva, E. V., Ananko, E. A., Podkolodnaya, O. A., Kolpakov, F. A., Podkolodny, N. L. and Kolchanov, N. A.: Databases on Transcriptional Regulation: TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res. 26, 362-367 (1998). (PubMed: 9399875)
3 Heinemeyer, T., Chen, X., Karas, H., Kel, A. E., Kel, O. V., Liebich, I., Meinhardt, T., Reuter, I., Schacherer, F. and Wingender, E.: Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 27, 318-322 (1999). (PubMed: 9847216)
4 Wingender, E.: Classification scheme of eukaryotic transcription factors. Molekularnaya Biologiya 31, 584-600 (1997); Mol. Biol. Engl. Tr. 31, 483-497 (1997). (PubMed: 9340487)
5 Pickert, L., Reuter, I., Klawonn, F. and Wingender, E.: Transcription regulatory region analysis using signal detection and fuzzy clustering. Bioinformatics 14, 244-251 (1998). (PubMed: 9614267)
GENUS (1993 – 1997)

The project "Gene regulatory nucleic acid sequences" (GENUS; funding code 01 IB 306 A) was one of the six projects that received financial support by the first pure bioinformatics funding program in Germany and, to my knowledge, world-wide. Working at the GBF at that time, I was lucky enough to coordinate this project which to join I could convince three excellent partners: Andreas Dress (University of Bielefeld), Heinz Sklenar (MDC, Berlin) and Thomas Werner (GSF, Oberschleißheim).
As the title of the project indicates, it was our aim to combine different computational, mathematical and biophysical approaches to characterize transcription factor binding sites, in order to come up with reliable prediction algorithms and tools for their recognition in genomic sequences. The goal of my team was to transform my transcription factor compilation1 into a real, relational database2, 3, 4 so that the collected information about TF binding sites could be used for a descriptive model of the DNA-binding specificity of as many transcription factors as possible.
As a result of this consortial research, the TRANSFAC database about transcription factors, their genomic binding sites and DNA-binding profiles was established and provided to the scientific community as a web-based resource. The attached library of positional scoring matrices was used by a tool, MatInspector, that we helped develop together with the group of Thomas Werner5. Heinz Sklenar came up with a comprehensive library of DNA-structural parameters, and together with him we could reveal some interesting structure-function relations for certain TF-DNA interactions6. One of the "comrade-in-arms" of these days, Holger Karas, has later joined me when founding the company BIOBASE and became its first CEO.
The final report of the whole consortial project from March 1997 can be found here (in German only).
Publications:
1 Wingender, E.: Compilation of transcription regulating proteins. Nucleic Acids Res. 16, 1879-1902 (1988). (PubMed: 3282223)
2 Knüppel, R., Dietze, P., Lehnberg, W., Frech, K. and Wingender, E.: TRANSFAC retrieval program: a network model database of eukaryotic transcription regulating sequences and proteins. J. Comput. Biol. 1, 191-198 (1994). (PubMed: 8790464)
3 Wingender, E., Dietze, P., Karas, H. and Knüppel, R.: TRANSFAC: A database on transcription factors and their DNA binding sites. Nucleic Acids Res. 24, 238-241 (1996). (PubMed: 8594589)
4 Wingender, E., Karas, H. and Knüppel, R.: TRANSFAC Database as a Bridge between Sequence Data Libraries and Biological Function. Pacific Symposium on Biocomputing '97 (PSB'97), R. B. Altman, A. K. Dunker, L. Hunter, T. E. Klein (eds.). World Scientific, Singapore - New Jersey - London - Hong Kong 1996, pp. 477-485. (PubMed: 9390316)
5 Quandt, K., Frech, K., Karas, H., Wingender, E. and Werner, T.: MatInd and MatInspector - New fast and sensitive tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23, 4878-4884 (1995). (PubMed: 8532532)
6 Karas, H., Knüppel, R., Schulz, W., Sklenar, H. and Wingender, E.: Combining structural analysis of DNA with search routines for detection of transcription regulatory elements. Comput. Appl. Biosci. 12, 441-446 (1996). (PubMed: 8996793)